Example Queries
- I need to design primers for my serpin gene in Tricuris muris but I want to check my gene model is correct based on my RNA-Seq data. Can I do this in WormBase ParaSite?
- I'd like to extract C. elegans orthologs for Nippostrongylus genes involved in a particular process.
- I have a list of genes from Ascaris suum and would like to know which ones have orthologs in humans and mammals and which ones might be nematode-specific:
- I need the sequences for a set of Schistosoma mansoni genes. I have the chromosome, start, and stop for each:
- I need a list of genes with predicted signal peptide that are present in Brugia malayia given organism but not present in C. elegans
I need to design primers for my serpin gene in Tricuris muris but I want to check my gene model is correct based on my RNA-Seq data. Can I do this in WormBase ParaSite?
-
Yes! First you will need to upload your RNASeq data into WormBase ParaSite.
See the Supported file formats section for more information on what format these files should be in.
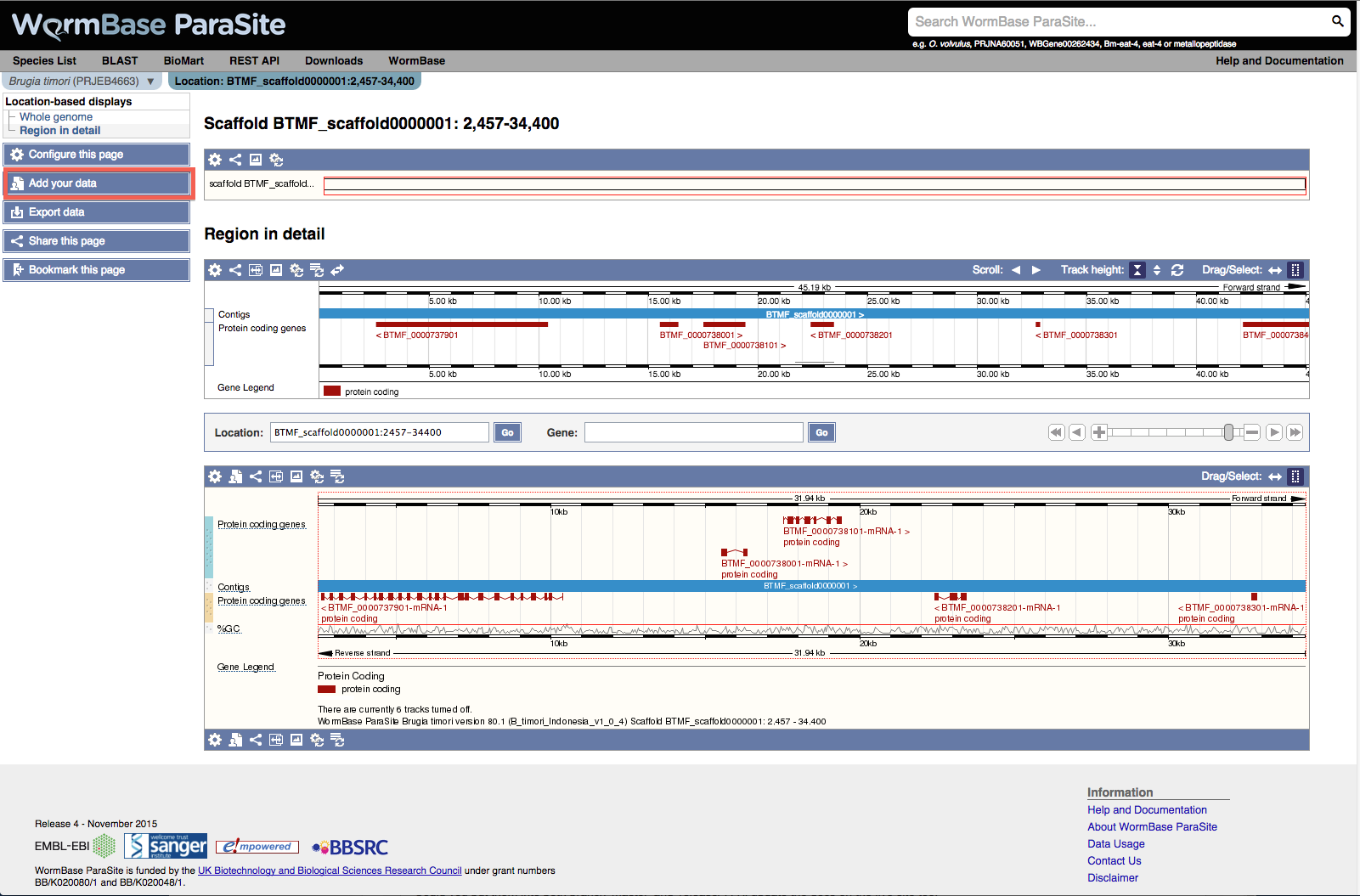
To add the data go to your region of interest for your species, for example.
- In the left panel, click Add custom tracks.
In the Data format pulldown, choose the correct format, in this case BigWig, and add the URL to your data, e.g.:
- Click Add data. Choose a colour for your track and click Save. You should now see your RNASeq data displayed as new tracks.
Add additional tracks using the same steps, or you can attach a group of tracks together if they are organised into a Track Hub.
The data will remain attached in your browser unless it is removed via the Configure tracks link. Tracks can be preserved between browsers/computers, and can also with collaborators by creating a WormBase ParaSite account.
- If you notice corrections that need to be made to gene models in WormBase ParaSite, please contact us.

-
I'd like to extract C. elegans orthologs for Nippostrongylus genes involved in a particular process.
- You can do that using BioMart. In the SPECIES menu select Nippostrongylus, then in the MULTI-SPECIES COMPARISONS menu select Orthologous C. elegans genes -> Only.
- You can further refine this list by function, process or location by choosing one or more categories from the GENE ONTOLOGY list. Start typing in the upper box and choose your terms of interest from the autocomplete, they will be added to the box beneath.
- Click the Results button, top left, to see your results. By default a two-column file is returned that contains gene ID and Genome Project. To configure different options for the output, select Attributes in the left menu. See the BioMart documentation for more information.

I have a list of genes from Ascaris suum and would like to know which ones have orthologs in humans and mammals and which ones might be nematode-specific:
ASU_00012 ASU_00034 ASU_00089 ASU_00299 ASU_00345 ASU_00890 ASU_00998 ASU_01289 ASU_03421 - You can do that using BioMart. In the GENE menu paste in your list, then in the MULTI-SPECIES COMPARISONS select Orthologous human genes -> Excluded.
You can also run this query against against mouse orthologs by selecting Orthologous mouse genes -> Excluded. The results are the same in this case.
- Click the Results button, top left, to see your results. By default a two-column file is returned that contains gene ID and Genome Project. To configure different options for the output, select Attributes in the left menu. See the BioMart documentation for more information.

I need the sequences for a set of Schistosoma mansoni genes. I have the chromosome, start, and stop for each:
- From the SPECIES filter choose Schistosoma mansoni. Open the REGION section and enter the list of co-ordinates under 'Multiple regions'. The list can be separated by commas or new lines.
- Click the Attributes option from the left menu. Check the Sequences option, then in the SEQUENCES section choose Unspliced (genes). Click the Results button, top left.
I need a list of genes with predicted signal peptide that are present in Brugia malayia given organism but not present in C. elegans.
| Smp.Chr_3:5955668:5958652:1 |
| Smp.SC_0395:8998:9186:1 |
| Smp.Chr_1:32692191:32693861:1 |
| Smp.Chr_3:26054871:26055183:-1 |


- You can do that using BioMart. In the SPECIES section choose Brugia malayi, then in the MULTI-SPECIES COMPARISONS select Orthologous C. elegans genes -> Excluded.
- In the PROTEIN DOMAINS section check Limit to genes... and from the menu select with signal P protein features -> Only
- Click the Results button, top left, to see your results. By default a two-column file is returned that contains gene ID and Genome Project. To configure different options for the output, select Attributes in the left menu. See the BioMart documentation for more information.