Gene Pages
The gene page is the source of information relating to a gene. All gene pages have a header section which summarizes the key information about the gene:

This includes (from top to bottom): Gene name and gene id, a short gene description (where available), location of the gene, additional information about the gene including number of orthologs, gene type, annotation method and a table of all alternative transcripts of the gene.
The menu on the left-hand side allows you navigate through available data:
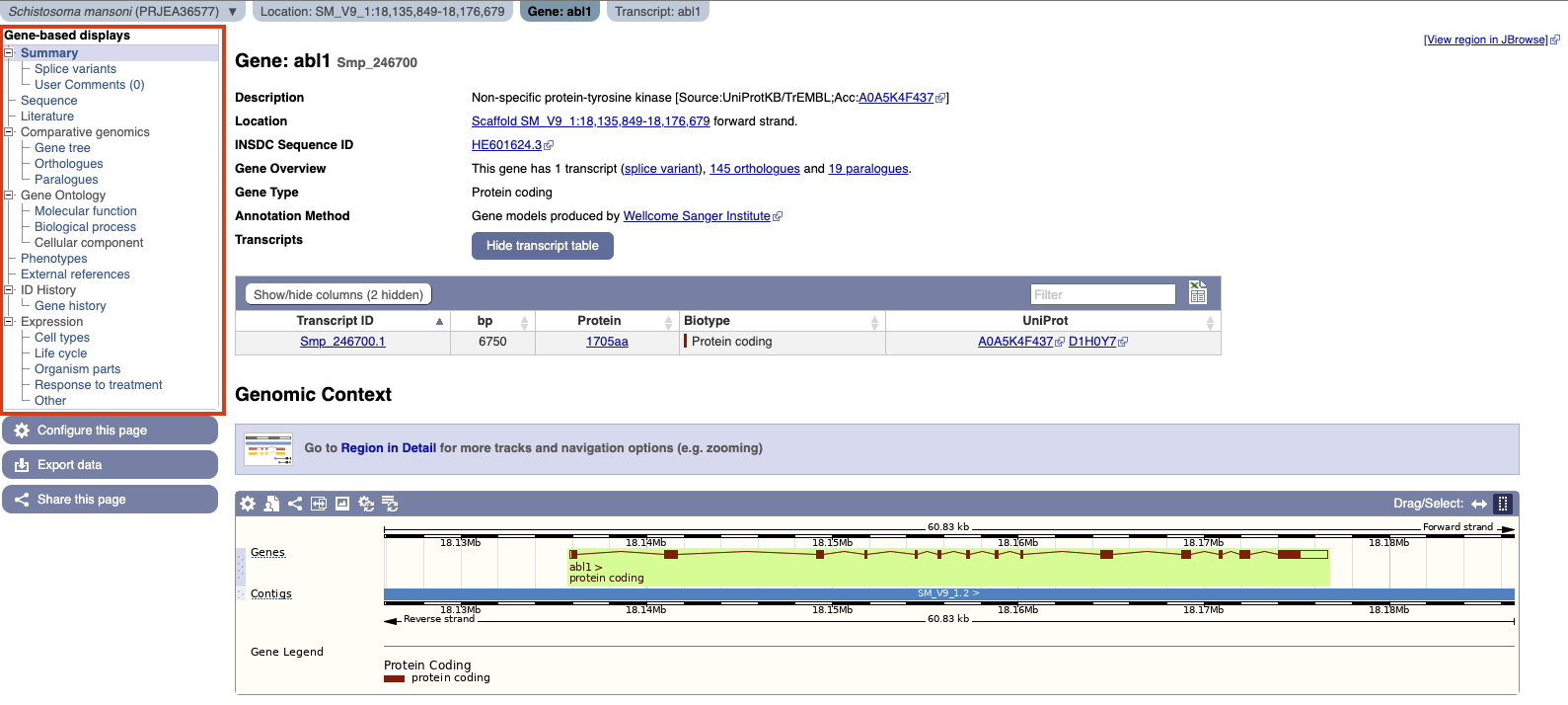
Available data are:
Summary
This is a summary view of the gene model, and any of its neighbours, on the genome. For a more interactive view, follow the Region in detail link.
Splice variants
Displays all known splice variants as a table and in a browser view.
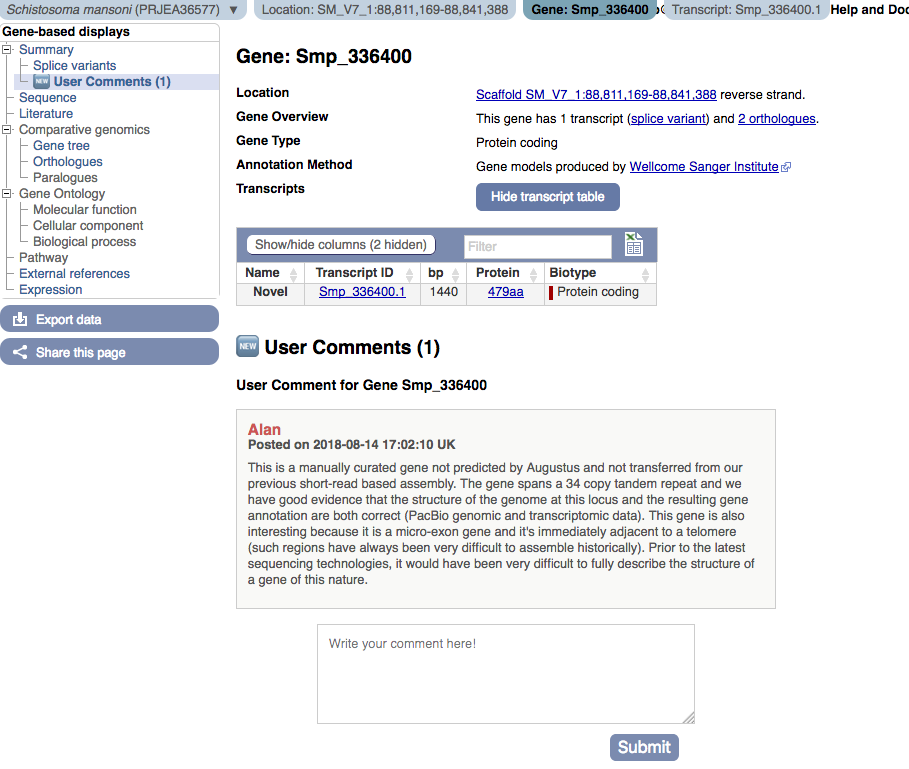
User comments
This is a space where anyone logged into their user account can add extra information about a given gene. Here, one of the authors of the annotation accompanying a Schistosoma mansoni Smansoni_v7 assembly remarks on location and exon structure of gene Smp_336400:
Sequence
This displays the nucleotide sequence of the gene. Exons are highlighted in pink/red:
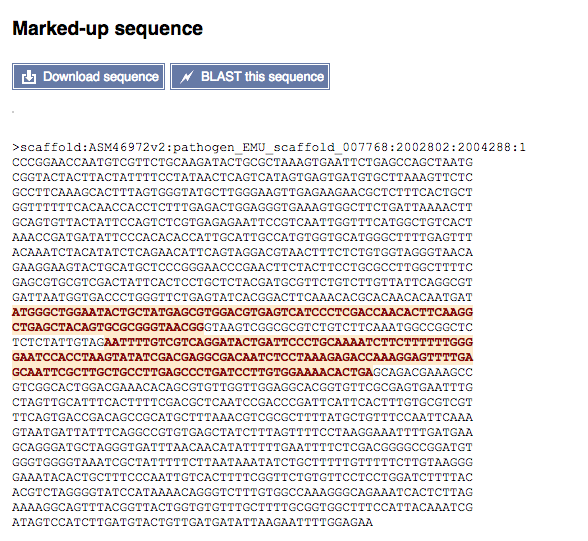
This page also gives you the option of downloading the sequence in FASTA or RTF formats, or to BLAST the sequence using the WormBase ParaSite BLAST service.
External references
Links to external databases such as EntrezGene and WormBase.
Literature
This page lists all papers from Europe PMC that mention this gene.
Phenotypes
This page lists all phenotypes associated with this gene or its orthologues in other species, along with the source/study supporting each association.
Gene Ontology
Displays all available Gene Ontology annotation data from the three ontologies: biological process, molecular function and cellular component. Clicking on the 'Chart' icon for a particular term displays a tree view of for that GO term up to the root node, and selecting the Ancestry chart tab shows a tree view for all annotated terms in the selected ontology.
Comparative Genomics
Gene tree
Find more about the Gene Tree page here.
Orthologues
Displays a list of all orthologous genes (i.e. evolved from a common ancestral gene) in species in WormBase ParaSite as well as some other comparitor and model species including Caenorhabiditis elegans, human, Drosophila melanogaster, Mouse and Zebrafish. The list can be filtered using the summary table at the top - by default all orthologs are displayed.
The alignment data underlying the table can be downloaded in a variety of different formats including FASTA, CLUSTAL W, Mega, MSF, Nexus, OrthoXML, Pfam, Phylip, PSI and Stockholm by following the link at the top of the table.
Paralogues
Displays a list of all paralogous genes (i.e. homologous gene sequences created by a duplication events within a genome).
The alignment data underlying the table can be downloaded in a variety of different formats including FASTA, CLUSTAL W, Mega, MSF, Nexus, OrthoXML, Pfam, Phylip, PSI and Stockholm by following the link at the top of the table.
Expression
Displays the Median Gene Expression (TPM values) for this gene across replicates of gene expression studies. Each subpage lists the gene expression values for a specific phenotype such as Cell types, Life cycle stages, Organism Parts, Response to treatment and more.