Genome Browser (Ensembl)
Our main genome browser - a tool enabling an interactive view of the genome - comes from Ensembl. It lets you view features of a chosen region, usually a selected gene.
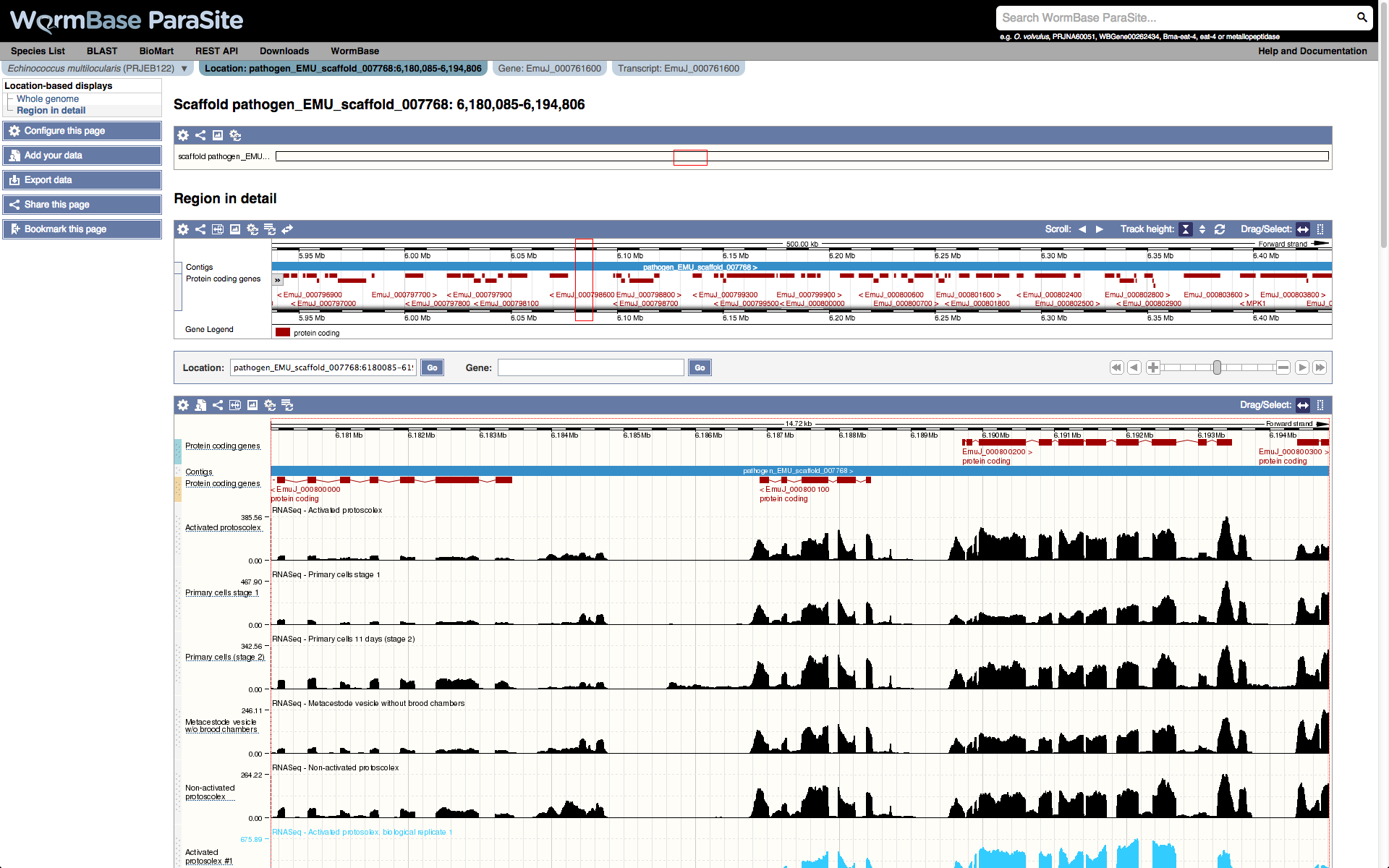
Whole scaffold/contig view
The top image shows the full scaffold or contig, with the location being displayed in detail highlighted by a red box. The selected area can be changed by clicking on the scaffold. This brings up a menu that allows the selected area to be re-centered or to zoom in or out. Alternatively you can select and drag to choose a new area of the scaffold or contig.
Region in detail
The second panel shows a zoomed-in view centered on the selected gene. The control panel underneath allows you to jump to a particular location or gene by entering them in each of the boxes on the left and clicking 'GO'.
On the right there is a control which allows you to scroll along the scaffold or contig by clicking the left or right arrows. The single arrow scrolls you along by a window width and the double arrow scrolls along 1MB. Alternatively, you can scroll by clicking and dragging the region in detail image. You can zoom in and out using the + and - , or by dragging the slider to the desired location.
Provided tracks
For all our genomes, we display gene models and a number of tertiary annotation features.
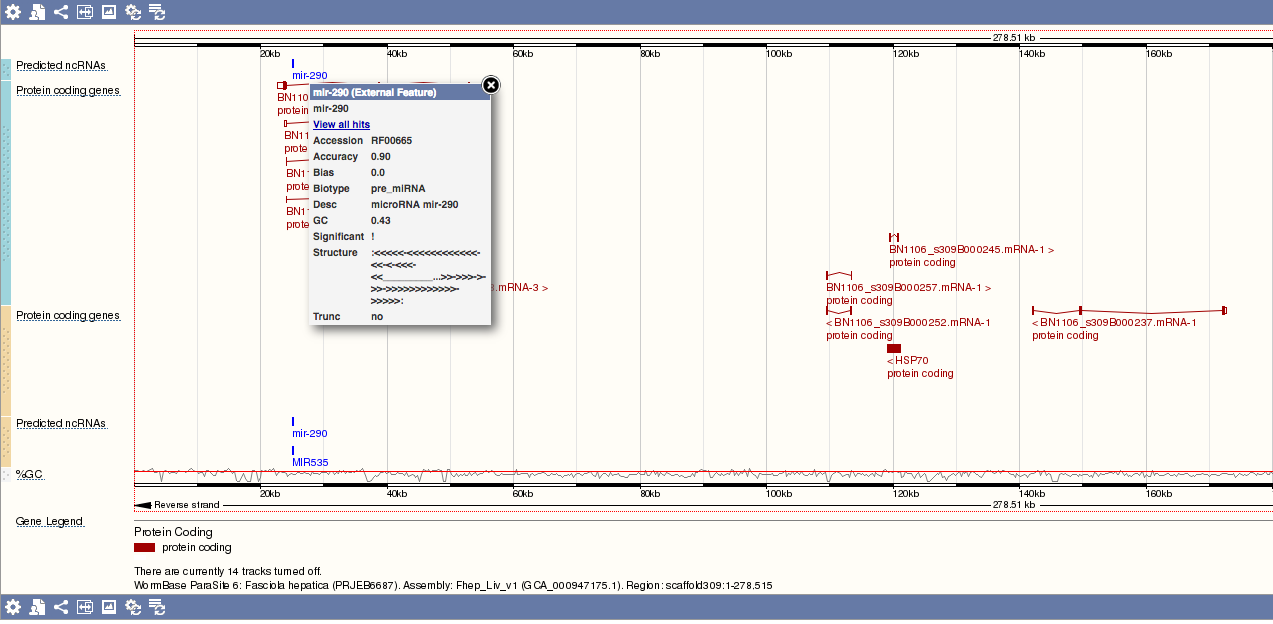
Predicted non-coding RNA (ncRNA)
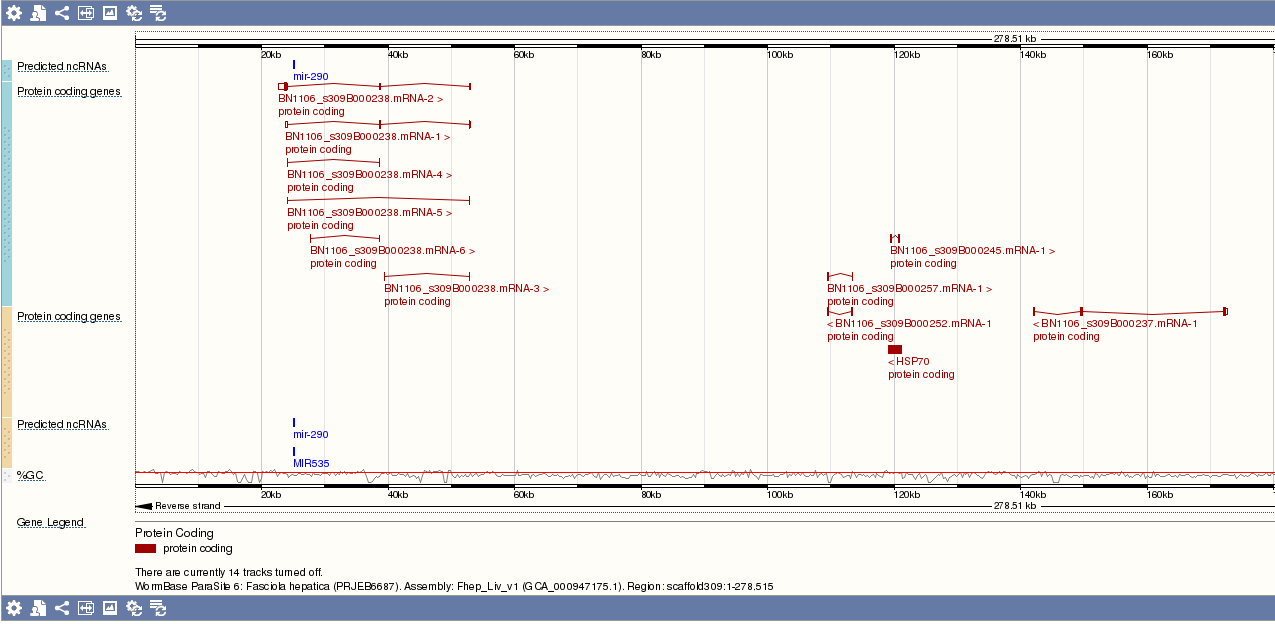
Clicking the ncRNA name brings up a window providing additional metadata about the ncRNA, and an option to view other hits on the genome:
RNASeq tracks
We collaborate with the RNASeq-er project ran by the Functional Genomics Group at the European Bioinformatics Institute.
RNASeq-er retrieves public RNASeq reads of Nematoda and Platyhelminthes deposited to European Nucleotide Archive and aligns them to WormBase ParaSite genomes. We import the alignment results, retrieve additional metadata, and add the tracks where available. The data is also viewable in our alternative genome browser based on JBrowse.
The tracks are sorted by ENA study accession:
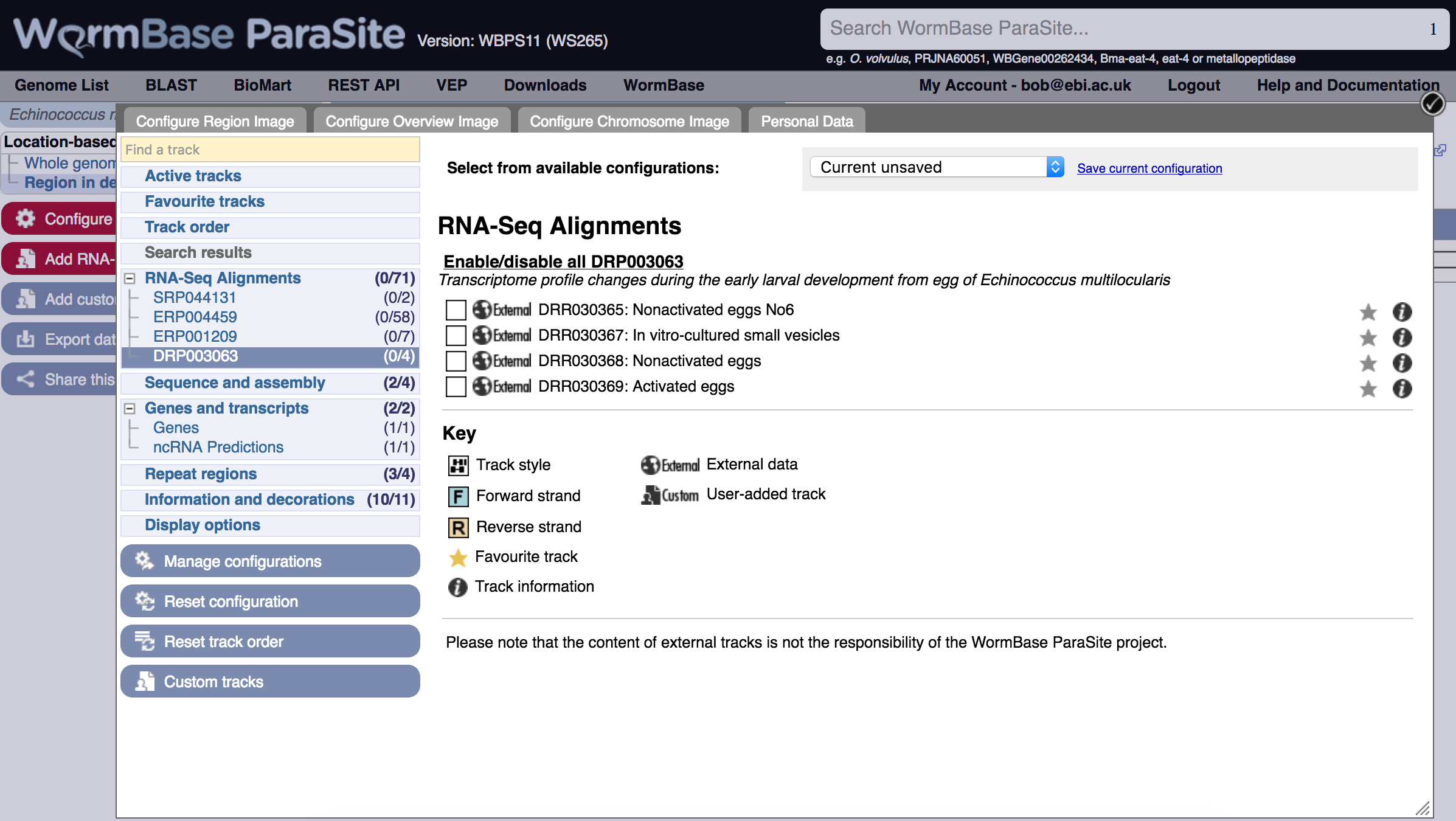
After selecting a track it appears in the genome view.
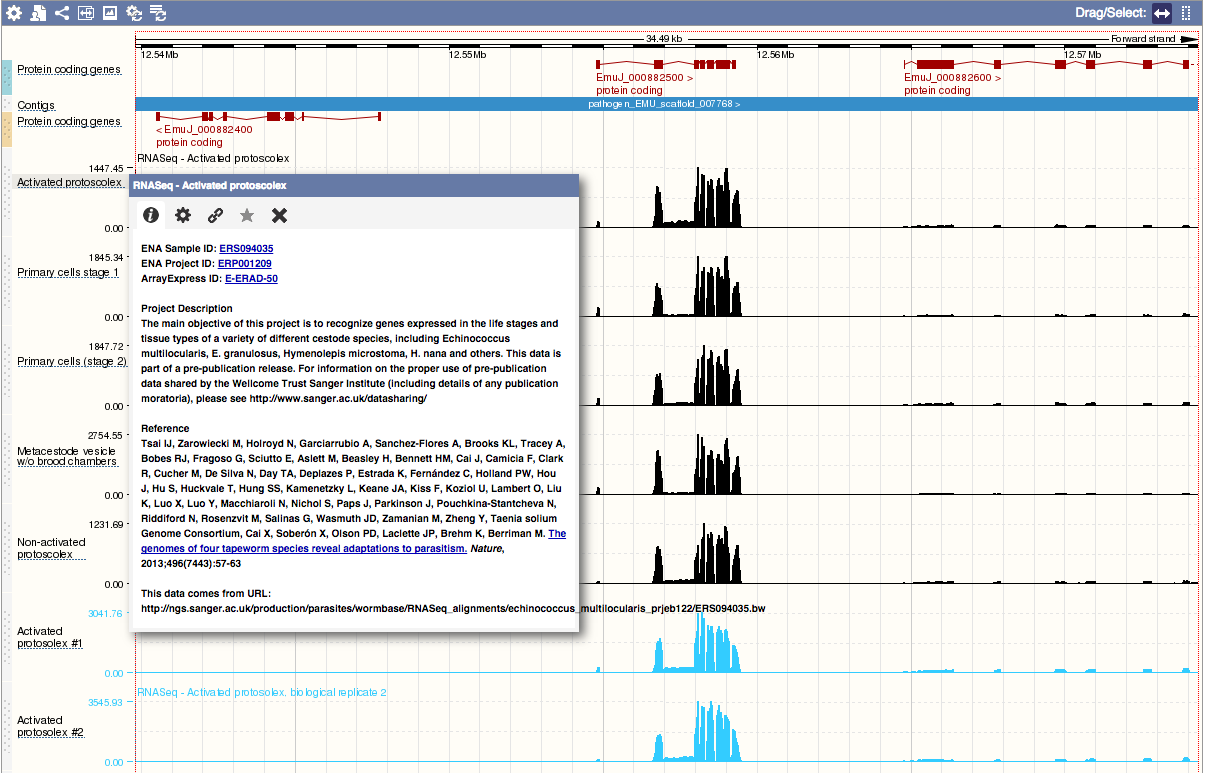
Hovering over the short track name to the left of the track brings up a window providing additional metadata about the track, and options to configure the track:
Hosted hubs
For some species - e.g. Ascaris suum (PRJNA62057) - we link externally provided hubs. Do contact us if you have a dataset that you think the community would consider valuable to include in this way.
Adding own tracks
You can add your own tracks to the display. Please see this page for more details.